Introduction
When studying diseases and medical interventions, patients often experience events that can occur multiple times (e.g., hospitalizations, infections, or tumor recurrences). Traditional survival analysis methods typically focus on time to the first event, potentially missing important information about the total disease burden.
Many patients experience events that can occur multiple times—like hospital readmissions, infections, or tumor recurrences. Traditional survival analysis is only concerned with the first event and does not account for competing risks like death. Alternatively, the mean cumulative count (MCC) provides a more complete picture by estimating the expected number of events per person by a given time, properly accounting for:
- Multiple occurrences of the same event (recurrent events)
- Events that prevent future occurrences (competing risks like death)
- Incomplete follow-up (censoring)
Clinical Example: Bladder Cancer Recurrences
We’ll use the survival::bladder1 dataset from the
survival package, which contains data on bladder cancer
recurrences from a randomized trial comparing three treatments:
placebo, pyridoxine, and
thiotepa.
The bladder1 dataset contains 118 patients followed for
tumor recurrences. The key variables are:
-
id: Patient identifier -
stop: Time of event or censoring -
status: Event type (0 = censored, 1 = recurrence, 2 = death from bladder disease, 3 = death from other cause) -
treatment: Treatment group (placebo, pyridoxine, or thiotepa)
Data Preparation
For MCC analysis, we need to ensure our competing event variable has the proper coding:
-
0= censoring -
1= event of interest (recurrence) -
2= competing event (death from any cause)
Basic MCC Analysis
Let’s calculate the MCC of bladder cancer recurrences for the entire cohort:
# Calculate MCC using the Dong-Yasui equation method
mcc_overall <- mcc(
data = bladder_mcc,
id_var = "id",
time_var = "stop",
cause_var = "status"
)
#> Warning: Found 13 participants where last observation is an event of interest
#> (`cause_var` = 1)
#> First 5 IDs: 13, 15, 16, 19, 24
#> Total affected: 13 participants
#> ℹ `mcc()` assumes these participants are censored at their final `time_var`
#> ℹ If participants were actually censored or experienced competing risks after
#> their last event, add those observations to ensure correct estimatesNotice that mcc() will warn the user if there are
participants whose data end do not explicitly have a censoring event
(0) of competing event (2) on their final row.
The function provides this warning to ensure the user is aware of this
and also aware of how this is implicitly interpreted by the package. The
function also lets the user know that the MCC estimate will be incorrect
if the mccount’s implicit assumption is incorrect.
Available Estimators
The mcc() function can be used to calculate the MCC
using either the Dong-Yasui estimator (method = "equation",
the default) or the sum of cumulative incidence estimator
(method = "sci"). See
vignette("choosing-between-methods") and ?mcc
for more details.
Understanding the Output
Just calling the mcc object we’ve created will print the
method used, a preview of the MCC estimates at each time point, and the
exact mcc() call used to generate the output:
mcc_overall
#>
#> ── Mean Cumulative Count Results ───────────────────────────────────────────────
#> ℹ Method: Dong-Yasui Equation Method
#>
#> ── MCC Estimates ──
#>
#> # A tibble: 6 × 2
#> time mcc
#> <int> <dbl>
#> 1 0 0
#> 2 1 0.0256
#> 3 2 0.112
#> 4 3 0.241
#> 5 4 0.276
#> 6 5 0.320
#> # ... with 45 more rows
#> ── Call ──
#>
#> mcc(data = bladder_mcc, id_var = "id", time_var = "stop", cause_var = "status")The package also includes an S3 method for the summary()
function that will output more useful details:
# Get summary statistics
summary(mcc_overall)
#>
#> ── Summary of Mean Cumulative Count Results ────────────────────────────────────
#> ℹ Method: Dong-Yasui Equation Method
#> ℹ Total participants: 118
#>
#> ── Summary Statistics ──
#>
#> Observation period: [0, 64]
#> Time to MCC = 1.0: 22
#> Time to maximum MCC: 53
#> MCC at end of follow-up: 2.2672
#>
#> ── Event Count Composition
#> Events of interest: 189
#> Competing risk events: 29
#> Censoring events: 76We can also extract the MCC estimate at the end of study follow-up
using mcc_final_values(mcc_overall) and/or extract the MCC
values at specific time points (not only the MCC at the end of
follow-up). For example:
# Extract key timepoints
key_times <- c(6, 12, 24, 36, 48, 60) # months
mcc_at_times <- mcc_overall$mcc_table |>
filter(time %in% key_times) |>
mutate(
years = time / 12,
mcc = cards::round5(mcc, 3)
) |>
select(years, time, mcc)
mcc_at_times
#> # A tibble: 6 × 3
#> years time mcc
#> <dbl> <int> <dbl>
#> 1 0.5 6 0.391
#> 2 1 12 0.622
#> 3 2 24 1.16
#> 4 3 36 1.64
#> 5 4 48 2.03
#> 6 5 60 2.27Visualization
mccount provides flexible plotting capabilities that can
be combined with ggplot2 functions/syntax:
# Basic plot
plot(mcc_overall) +
labs(
title = "Bladder Cancer Recurrence Burden",
subtitle = "Mean cumulative count over time",
x = "Time (Months)"
) +
scale_x_continuous(
breaks = c(seq(
0, max(mcc_overall$mcc_table$time),
by = 12
)
)) +
geom_line_mcc(mcc_overall)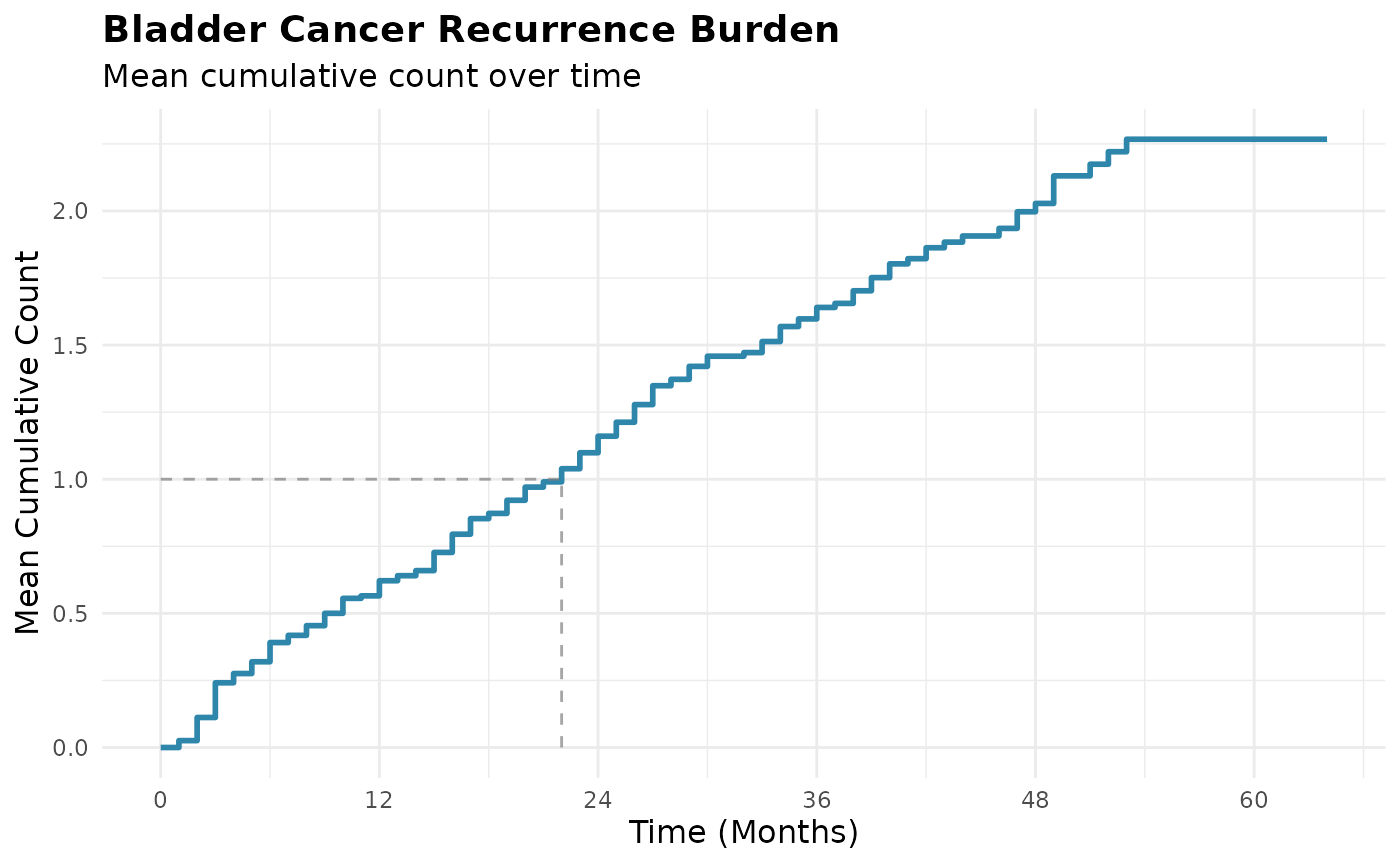
The step function shows how the expected number of bladder cancer recurrences accumulates over the entire study follow-up period.
MCC Interpretation
The estimated MCC can be interpreted as the number of events per
person we expect among individuals who have not experienced a competing
risk event by a given time. Looking at the print out from
summary(mcc_overall), the MCC at the end of follow-up is
2.27 - on average, each bladder cancer survivor experiences 2.27
recurrences by 64 months.
In a population of 100 similar patients, we expect approximately
2.27 * 100 = 227 total recurrences by 64 months. This
accounts for the fact that some patients die and cannot experience
further recurrences.
Dong, et al.1 provide additional guidance for interpreting the MCC (emphasis added):
Note that MCC is a marginal measure (as opposed to a conditional measure) of disease burden, similar to [cumulative incidence], and its interpretation is not conditional on survival free of competing risk events. Also, MCC does not assume independence between the event of interest and the competing-risk event.
Stratifying by Treatment Groups
It’s common that we may want to estimate the MCC stratified by a
specific characteristic, like "treatment" group.
mcc() provides a simple by argument to assist
with this:
# Calculate MCC by treatment group
mcc_by_treatment <- mcc(
data = bladder_mcc,
id_var = "id",
time_var = "stop",
cause_var = "status",
by = "treatment",
method = "equation"
)
#> Warning: Found 7 participants where last observation is an event of interest
#> (`cause_var` = 1)
#> First 5 IDs: 13, 15, 16, 19, 24
#> Total affected: 7 participants
#> ℹ `mcc()` assumes these participants are censored at their final `time_var`
#> ℹ If participants were actually censored or experienced competing risks after
#> their last event, add those observations to ensure correct estimates
#> Warning: Found 4 participants where last observation is an event of interest
#> (`cause_var` = 1)
#> ! ID: 51, 64, 67, 70
#> ℹ `mcc()` assumes these participants are censored at their final `time_var`
#> ℹ If participants were actually censored or experienced competing risks after
#> their last event, add those observations to ensure correct estimates
#> Warning: Found 2 participants where last observation is an event of interest
#> (`cause_var` = 1)
#> ! ID: 83, 104
#> ℹ `mcc()` assumes these participants are censored at their final `time_var`
#> ℹ If participants were actually censored or experienced competing risks after
#> their last event, add those observations to ensure correct estimates
# Summary by group
summary(mcc_by_treatment)
#>
#> ── Summary of Mean Cumulative Count Results ────────────────────────────────────
#> ℹ Method: Dong-Yasui Equation Method
#> ℹ Total participants: 118
#> ℹ Overall observation period: [0, 64]
#>
#> ── Summary by Group (treatment) ──
#>
#> ── Group: placebo
#> Participants in group: 48
#> Group observation period: [0, 64]
#> Time to MCC = 1.0: 17
#> Time to maximum MCC: 53
#> MCC at end of follow-up: 2.6137
#> Events of interest: 87
#> Competing risk events: 11
#> Censoring events: 30
#>
#> ── Group: pyridoxine
#> Participants in group: 32
#> Group observation period: [0, 60]
#> Time to MCC = 1.0: 19
#> Time to maximum MCC: 49
#> MCC at end of follow-up: 2.5831
#> Events of interest: 57
#> Competing risk events: 7
#> Censoring events: 21
#>
#> ── Group: thiotepa
#> Participants in group: 38
#> Group observation period: [0, 59]
#> Time to MCC = 1.0: 27
#> Time to maximum MCC: 47
#> MCC at end of follow-up: 1.5463
#> Events of interest: 45
#> Competing risk events: 11
#> Censoring events: 25Visualizing Treatment Differences
The other functions in mccount automatically recognize
the mcc_grouped S3 class and adjust their behavior
accordingly. For example, we can feed mcc_by_treatment into
plot(), and it automatically knows to plot an MCC curve by
the stratification variable.
# Plot with treatment groups
plot(mcc_by_treatment) +
labs(
title = "Bladder Cancer Recurrence Burden by Treatment",
subtitle = "Mean cumulative count over time",
x = "Time (Months)",
color = "Treatment"
) +
scale_x_continuous(
breaks = seq(
0, max(mcc_by_treatment$original_data$time),
by = 12
)
) +
geom_line_mcc(mcc_by_treatment)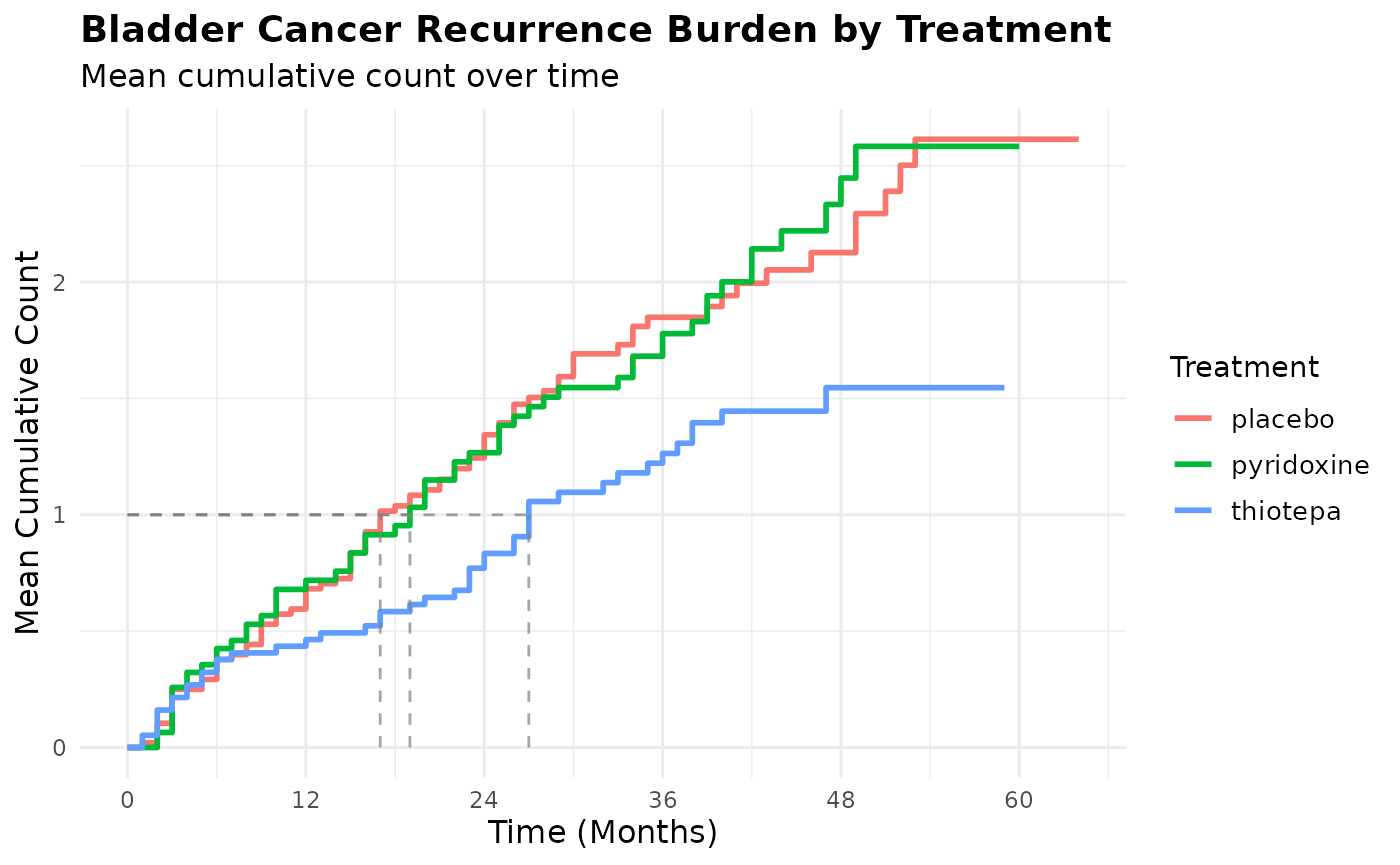
Confidence intervals for the MCC can be generated via bootstrapping.
Weighted MCC Analysis
mccount also allows estimation of weighted MCC in the
setting of observational studies that want to account for measured
confounding and/or other types of bias via matching or weighting. See
the vignette("estimating-mcc-after-matching-or-weighting")
for more details.